Proteins often consist of hundreds or thousands of individual parts, the amino acids. These are connected like the links of a chain. However, protein molecules would not be able to fulfill their task as a long filament flapping back and forth. Each of them is therefore folded in its own, very specific way during its creation. For proteins that are released from the cell to the outside or transported to intracellular stores, this folding takes place at a specific location in the cell: the endoplasmic reticulum (ER). Here, the partial strands that end up lying next to each other in the course of this protein folding can also be linked together at defined points. This prevents the ball from unraveling and retains its functional shape.
The connection is made via a mechanism that many people are already familiar with in a completely different context - in the hair salon. That is because it is used in perming to keep the hair in shape. For this purpose, certain chemicals are applied to the curlers. They ensure that chemical bonds, known as disulfide bridges, are formed between neighboring hair proteins. "Disulfide bridges, which link the neighboring sub-strands, are also formed at certain points in the protein filament during protein folding," explains Prof. Dr. Andreas Meyer of the Institute of Crop Science and Resource Conservation (INRES) at the University of Bonn.
Molecular "bucket line" passes electrons onward
Chemically, this process is an oxidation: This is because certain amino acids contain bound hydrogen sulfide (chem: S-H). If two such amino acids come to lie next to each other during the folding of the filament and one splits off the hydrogen (H) from their S-H groups, a disulfide bridge (chem: S-S) forms. For this process, one electron is removed from each of the two original S-H bonds. This is done by certain proteins that exert a precisely matching "traction force" on the electrons. However, the removed electrons must be guided out of this environment, otherwise they can cause damage.
When there is a fire, people sometimes form a bucket line, passing the full bucket of water from one person to the next. There is something similar in plants: The removed electrons are passed on through a chain of supporting proteins and finally disposed of. For this to work, each of the proteins involved along this chain needs to have a little more traction - in research, this is also referred to as redox potential. "These processes are in principle understood mechanistically," Meyer explains. "However, exactly how large the redox potentials are in living cells and how the entire reaction chain works dynamically is unknown."
But the degree of traction is critical for protein folding to produce the desired result: If it is too strong, too many S-H groups are oxidized. The amino acid ball is then linked in the wrong places and does not take the correct shape. In contrast, a too low redox potential results in insufficient stabilization because too few disulfide bridges are formed.
"We have now developed a method to measure the redox potential of one of the molecules involved in the ER of the living cell," says Meyer's collaborator José Manuel Ugalde. "Since the redox potential can change over time, we do this at three-minute intervals over a period of several hours." The measurement is made with a sensor protein that emits light itself, i.e. fluoresces, when irradiated with a laser. The emitted light intensity depends on the redox potential of the measured molecule. The researchers focused on the last link in the "bucket line," the ER oxidoreductins (EROs). These transfer the electrons snatched from the proteins to oxygen, thereby disposing of them.
Less traction than expected
The groups from Münster and Bonn used Arabidopsis thaliana as an experimental plant. It has two slightly different EROs whose building instructions are on two different genes. "We were able to show that the EROs exert a much lower traction force on electrons than previously thought," Meyer explains. "So the whole chain is probably calibrated differently."
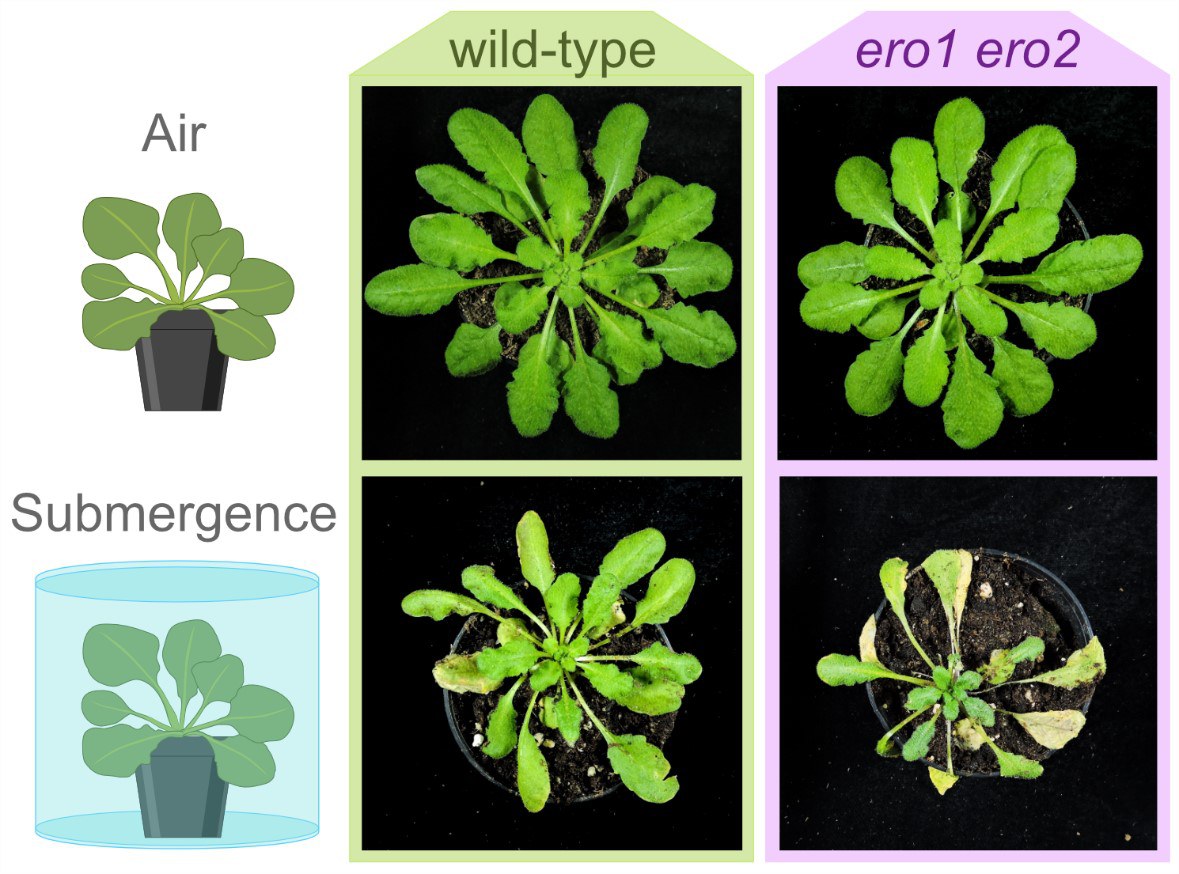
The EROs need oxygen to get rid of their electrons. However, when waterlogging occurs - for example, due to excessive watering of potted plants or after a flood in the field - oxygen is scarce for parts of the plants. Under these conditions, the EROs appear to adjust the redox potentials in the bucket line in a way that allows disulfide bridge formation to still proceed, albeit at a reduced rate. This means that the plant can grow even in the absence of oxygen. To study this effect in more detail, the researchers switched off one of the ERO genes completely and made the second one significantly more inactive. As a result, the plant was much more sensitive to oxygen deficiency and other types of reductive stress. For example, it then grew only very slowly. "Nevertheless, protein folding still worked, at least partially, under these conditions," says Meyer, who is also a member of the Transdisciplinary Research Area "Sustainable Futures" at the University of Bonn. "We therefore hypothesize that there is a second mechanism in Arabidopsis that accomplishes the oxidation of S-H residues in proteins in the ER."
The researchers now want to find out exactly which one. In the long term, their findings could therefore potentially contribute to the development of plant varieties that are better able to withstand short-term oxygen deprivation. With climate change increasing not only droughts but also floods, demand for these is likely to rise significantly in the future.
Participating institutions and funding:
In addition to the University of Bonn, the Universities of Münster and Bielefeld and the Technical University of Kaiserslautern were involved in the study. The work was funded by the German Research Foundation (DFG).
Publication:
José Manuel Ugalde, Isabel Aller, Lika Kudrjasova, Romy R. Schmidt, Michelle Schlößer, Maria Homagk, Philippe Fuchs, Sophie Lichtenauer, Markus Schwarzländer, Stefanie J. Müller-Schüssele, Andreas J. Meyer: Endoplasmic reticulum oxidoreductin provides resilience against reductive stress and hypoxic conditions by mediating luminal redox dynamics. Plant Cell; https://doi.org/10.1093/plcell/koac2021